
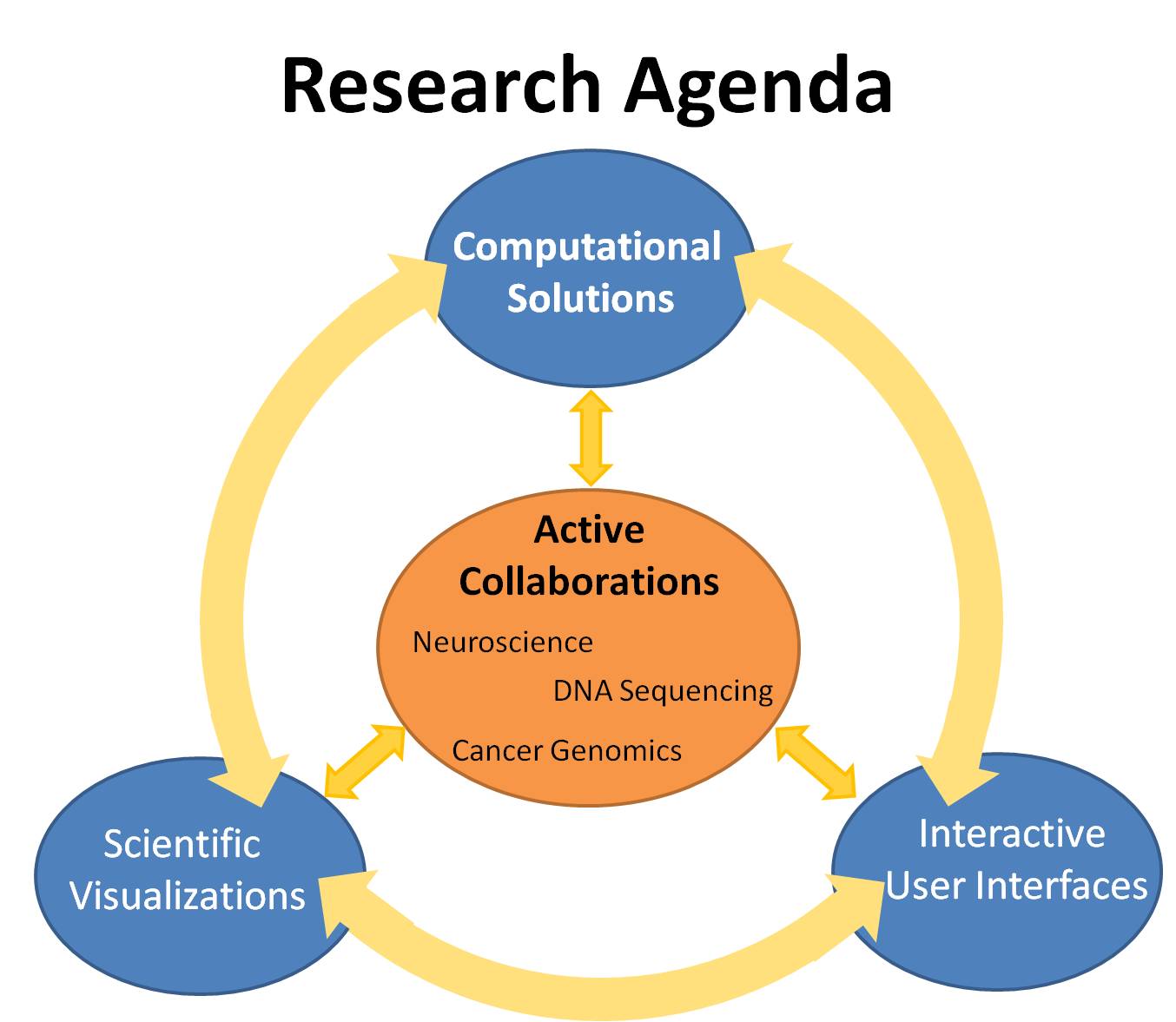
The diagram above outlines the three research areas that I work on. The first is the computational solutions for extraction of "hidden'' information and physical properties in medical imaging data from various scientific disciplines through mathematical modeling and analysis. The second area explores powerful visualization tools for combining multi-dimensional information obtained from various imaging modalities. The third area attacks the problem of designing efficient interactive user interfaces for understanding and exploring scientific data at multiple scales. These three research areas interact with one another to expand our knowledge in computer science, neuroscience and life sciences.
In particular, I have been focusing on developing computational techniques and interactive visualization applications through close collaboration with neuropsychologists, including Dr. Stephen Correia at Butler Hospital and Dr. David Tate at Harvard Medical School, to improve understanding of the brain and solve medical problems. I have developed new computational approaches towards the virtual histology of brain microstructure using diffusion imaging to provide reliable and sensitive biomarkers for brain disease diagnosis and prevention. The information extraction aspect of the work is meaningful in many other health-related areas of life sciences such as biology, DNA sequencing and cancer genomics, which I plan to explore. I have also designed interactive visualization interfaces for exploring multidimensional brain data in order to help brain scientists test their hypothesis on brain segmentation, connectivity and pathological changes. I aim to investigate the design of powerful visualizations that encode multidimensional information and efficient interactive interfaces that explores multiscale data across various scientific domains.
- Part I: Quantitative Computational Modeling and Data Analysis
- Part II: Qualitative Visualization
- Part III: Interactive User Interfaces
I. Quantitative Microstructure Measurements: Virtual Histology
Motivation: The microstructural properties of brain white matter, such as axon radius, directly affect the brain's nerve function and have been observed to correlate with various neurological diseases like multiple sclerosis and tumor-grading. Despite the importance of these microstructural properties, they have not been reliably measurable in vivo, so that their investigation has mainly relied on invasive histology examinations. These examinations in traditional neuroanatomy often introduce artifacts and cannot be used to analyze disease stages and monitor progression.
The goal here is to replace this invasive histology examination with a non-invasive in-vivo computational approach to extract specific microstructural measures of underlying tissue properties that can provide reliable and sensitive biomarkers for disease-induced changes using diffusion MRI.
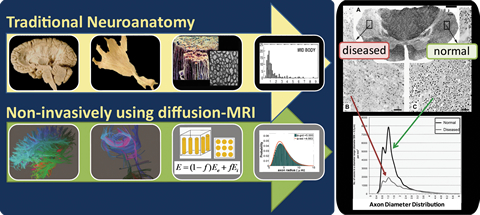
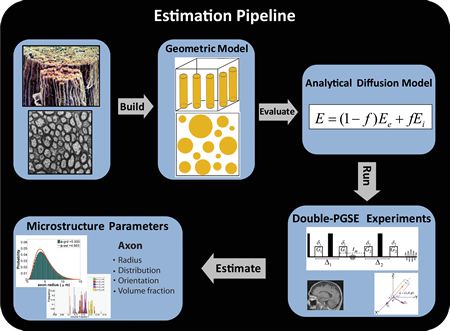
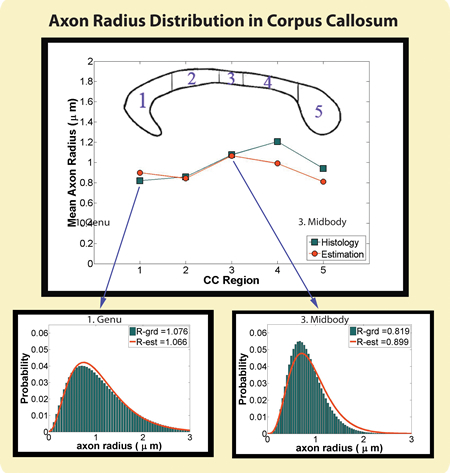
Results: With my new computational approaches through analytical modeling and analysis, I was able to extract specific microstructural measures of the underlying tissue including axon sizes, distribution and volume fraction. I thus obviated some of the limitations of earlier work by (1) improving sensitivity to small axons (radius < 3 micrometer) that are more vulnerable to disease and studying the full range of axon sizes in the human brain, (2) providing more realistic model that takes into account axon radii variation, (3) making the approach applicable to low-gradient clinical scanners and whole-brain mapping by mathematically modeling the water diffusion using a new diffusion MRI protocol. My results have been presented at various peer-reviewed international conferences and my recent submission to NeuroImage is a complete validation study on the feasibility and reliability of these computational approaches. My future work in brain science will build more sophisticated computational models in order to extract other tissue compartments including neurons, glial cells and dendrites as well as microstructural properties of gray matter.
Related Publications:
- DoubleAx: estimating axonal properties using angular double-pulsed gradient-spin-echo MRI in tissue of unknown orientation. Wenjin Zhou, Matt G. Hal and David H. Laidlaw. NeuroImage , 2001. (In review)
- Measurement of axon radii distribution in orientationally unknown tissue using angular double-pulsed gradient spin echo (double-PGSE) NMR. Wenjin Zhou and David H. Laidlaw. In International Society for Magnetic Resonance in Medicine - ISMRM , 19:3938, 2011. (Montreal, QC, Canada)
- Inferring Microstructural Properties Using Angular Double Pulsed Gradient Spin Echo NMR in Orientationally Unknown Tissue. Wenjin Zhou, Matt G. Hall, and David H. Laidlaw. In computational Diffusion MRI (CDMRI) Workshop at International Conference on Medical Image Computing and Computer-Assisted Intervention - MICCAI, 2010. (Beijing, China)
- Inferring Axon Properties with double-PGSE MRI using Analytical Water Diffusion Model. Wenjin Zhou and David H. Laidlaw. In International Society for Magnetic Resonance in Medicine - ISMRM, chosen for podium talk, 18:677, 2010. (Stockholm, Sweden)
- An Analytical Model of Diffusion and Exchange of Water in White Matter from Diffusion-MRI and its Application in Measuring Axon Radii. Wenjin Zhou and David H. Laidlaw. In International Society for Magnetic Resonance in Medicine - ISMRM, chosen for podium talk, 17:263, 2009. (Hawaii, USA)
II. Qualitative Visualization
Motivation: Visualize brain white-matter spatial relationships based on similarity measures from diffusion MRI for understanding brain connectivity.
Results: I demonstrated the benefits of visualization in disentangling the complex neural tractography data and in helping brain scientists understand and identify the underlying neuroanatomy and its connectivity. My ACM SIGGRAPH 2006 BEST POSTER which won FIRST PLACE in the ACM student research competition demonstrated a smooth-perceptual coloring scheme to map similar tracts to similar colors in order to help the users visually identify meaningful anatomical structures without imposing a rigid segmentation. In my later work in VIS 2007, I mapped this similarity measure into stripe texture patterns and improved the visualization of subtle differences within large white-matter structures.

Motivation: Visualize cortical-connectivity integrity and pathological changes
Results: A circular visualization interface of the tractography metrics for assessing cortical-connectivity integrity. The outer ring represents the 80 subdivided grey matter regions-of-interest (ROIs); the intensity of the edge corresponds to the normalized value of the tractography metrics across all subjects. The visualization allowed users to analyze various tractography metrics and identify the location of cortical-connectivity difference between healthy control and patient groups.
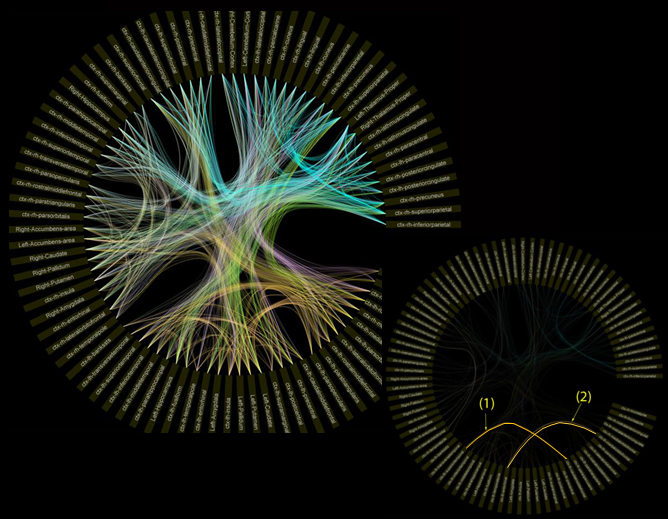
Related Publications:
- Visualizing tractography metrics of cortical-connectivity integrity in diffusion imaging. .Radu Jianu, Wenjin Zhou, Ryan Cabeen, Daniel Dickstein, and David H. Laidlaw. In International Society for Magnetic Resonance in Medicine - ISMRM ,20:3614, 2012. (Melbourne, Australia)
- Visualizing Spatial Relations Between 3D-DTI Integral Curves Using Texture Patterns, Doria Jianu, Wenjin Zhou, Cagatay Demiralp, and David H. Laidlaw, in IEEE Visualization Poster Compendium, 2007. (Sacramento, CA, USA)
- Perceptual Coloring and 2D Sketching for Segmentation of Neural Pathways. Wenjin Zhou, Peter G. Sibley, Song Zhang, David F. Tate, and David H. Laidlaw, In SIGGRAPH Poster Session, 2006. (Boston, MA, USA)
- BEST POSTER in SIGGRAPH 2006
- First place in ACM Student Research Competitions
- Abstract
- Poster
- Pictures
III. Interactive User Interfaces
Motivation: Segmentation of white-matter pathways by interactively selecting tracts-of-interest (TOI) has become a popular way for brain scientists to test their hypotheses on white-matter connectivity and quantitative pathological analysis.
Results: I conducted evaluations and developed taxonomy and design guidelines for TOI selection as a framework for exploring and categorizing the design space of the techniques in order to analyze their utility, usability, accuracy and reliability. Good user-interfaces should be user- and data-oriented. I designed a 2D sketching interface to match the design interface to neuroscientists' training in identifying anatomical structures on 2D axis-aligned slices. The interface reduced the high-dimensional data to a 2D projection that neuroscientists can easily understand. I later extended the 2D lasso-drawing selection mechanism to 3D stereo virtual reality (VR) environment with higher-input devices for better manipulation of 3D data. I presented my results as one of the top five BEST POSTER nominees at VIS 2008. The VR environment in this interface reduced visual clutter; the higher-order input device greatly reduced navigation time (a key challenge in TOI selection tools) and especially selection time for these tortuous structures.

Related Publications:
- Quantitative diffusion tensor imaging tractography metrics are associated with cognitive performance among HIV-infected patients. David F. Tate, Jared Conley, Robert H. Paul, Kathryn Coop, Song Zhang, Wenjin Zhou, David H. Laidlaw, Lynn E. Taylor, Timothy Flanigan, Bradford Navia, Ronald Cohen and Karen Tashima. Brain Imaging and Behavior, 4:68-79, 2010.
- Haptics-Assisted 3D Lasso Drawing for Tracts-of-interest Selection in DTI Visualization. Wenjin Zhou, Stephen Correia, and David H. Laidlaw. In IEEE Visualization Poster Compendium, 2008. (Columbus, OH, USA)
- Evaluation of Design Features in Interactive 3D Tracts-of-interest Selection Tools in DTI. Wenjin Zhou, Stephen Correia, and David H. Laidlaw. In IEEE Visualization Poster Compendium, 2008. (Columbus, OH, USA)
- Perceptual Coloring and 2D Sketching for Segmentation of Neural Pathways. Wenjin Zhou, Peter G. Sibley, Song Zhang, David F. Tate, and David H. Laidlaw, In SIGGRAPH Poster Session, 2006. (Boston, MA, USA)
- BEST POSTER in SIGGRAPH 2006
- First place in ACM Student Research Competitions
- Abstract
- Poster
- Pictures